Making the Impossible Possible with Tachyon: Accelerate Spark Jobs from Hours to Seconds
Cluster computing and Big Data technologies have enabled analysis on and insights into data. For example, a big data application might process data in HDFS, a disk-based, distributed file system. However, there are many reasons to avoid storing your on data disk, such as for data regulations, or for reducing latency. Therefore, if you need to avoid disk read/writes, you can use Spark to process the data, and temporarily cache the results in memory.
There are a number of use cases where you might want to avoid storing your data on disk in a cluster, in which case our configuration of Tachyon makes this data available in-memory in the long-term and shared among multiple applications.
However, in our environment at Barclays, our data is not in HDFS, but rather, in a conventional relational database management system (RDBMS). Therefore, we have developed an efficient workflow in Spark for directly reading from an RDBMS (through a JDBC driver) and holding this data in memory as a type-safe RDD (type safety is a critical requirement of production-quality Big Data applications). Since the database schema is not well documented, we read the raw data into a dynamically-typed Spark DataFrame, then analyze the data structure and content, and finally cast it into an RDD. But there is a problem with this approach.
Because the data sets are large, it can take a long time to load from an RDBMS, so loading should be done infrequently. Spark can cache the DataFrame in memory, but the cached data in Spark is volatile. If we have to restart the Spark context (for example due to an error in the code, null exceptions or changes to the mapping logic) we will then have to reload the data, which could take (in our case) half an hour or more of downtime. It is not unusual to have to do this a number of times a day. Even after we have successfully defined the mapping into typed case classes, we still have to re-load the data every single time we run a Spark job, for example if there is a new feature we want to compute, a change in the model, or a new evaluation test.
We need an in-memory storage solution.
Tachyon is the in-memory storage solution. Tachyon is the in-memory storage layer for data, so any Spark application can access the data in a straightforward way through the standard file system API as you would for HDFS. Tachyon enables us to do transformations and explorations on large datasets in memory, while enjoying the simple integration with our existing applications.
In this article, we first present how our existing infrastructure loads raw data from an RDBMS and uses Spark to transform it into a typed RDD collection. Then, we discuss the issues we face with our existing methodology. Next, we show how we deploy Tachyon and how Tachyon greatly improves the workflow by providing the desired in-memory storage and minimizing the loading time at each iteration. Finally, we discuss some future improvements to the overall architecture.
Previous Architecture
Since the announcement of DataFrame in Spark 1.3.0 (experimental) and its evolution in recent releases (1.5.0+), the process of loading any source of data has become simple and nicely abstracted.
In our case, we generate parallel JDBC connections which partition and load a relational table into a DataFrame. The DataFrame rows are then mapped into case classes.
Our methodology allows us to process raw data directly from source and build our code even though the data is not physically available to the cluster disks.
The following is our typical iterative workflow:
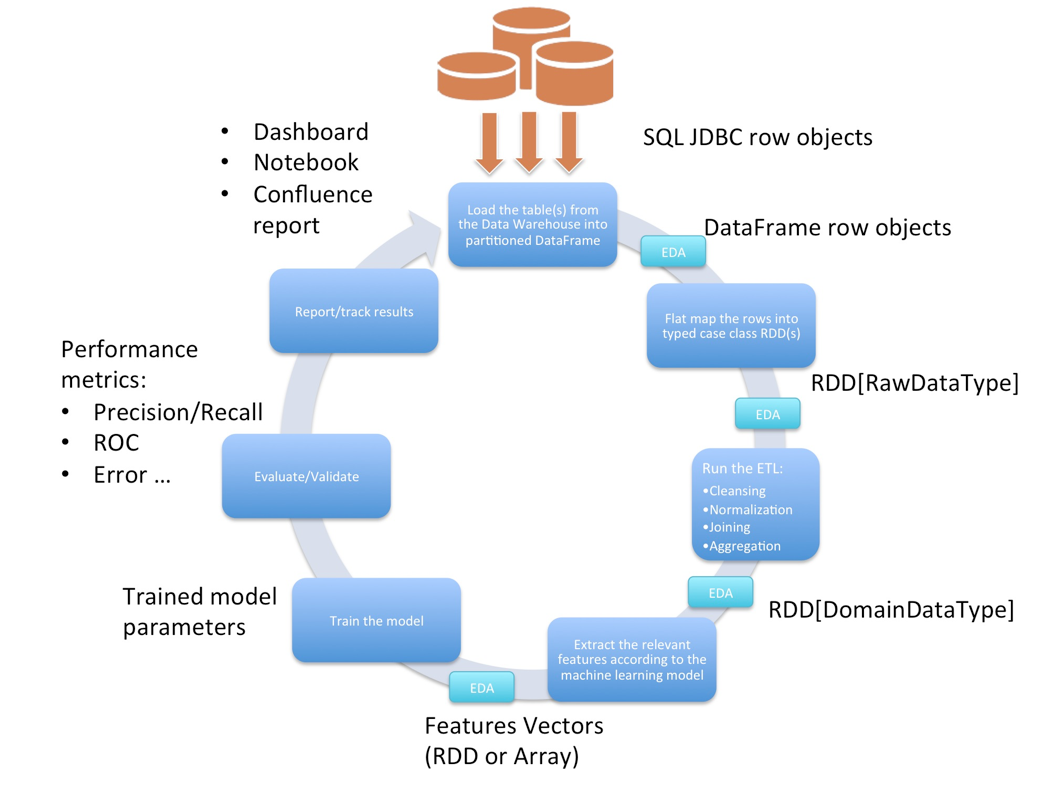
Setting up the JDBC Drivers
In order to create a JDBC source DataFrame, you must distribute the JDBC drivers jar (or jars) to each node of your cluster. Please ensure that the drivers must be available when you instantiate the JVM for your job. You cannot simply specify those jars as you would for normal dependencies even though it might be possible to distribute them using resource managers such as YARN.
We use a script to copy those files to the local file system of each node and then submit the Spark job by specifying both the executor and driver extraClassPath properties. Make sure that spark.driver.extraClassPath does not work in client-mode. Spark documentation says:
Note: In client mode, this config must not be set through the SparkConf directly in your application, because the driver JVM has already started at that point. Instead, please set this through the --driver-class-path command line option or in your default properties file.
Thus the following is an example of properly setting the drivers:
--driver-class-path "driver_local_file_system_jdbc_driver1.jar:driver_local_file_system_jdbc_driver2.jar"
--class "spark.executor.extraClassPath=executors_local_file_system_jdbc_driver1.jar:executors_local_file_system_jdbc_driver2.jar"The JDBC driver class must be visible to the primordial class loader on the client session and on all executors. This is because Java’s DriverManager class does a security check that results in it ignoring all drivers not visible to the primordial class loader when one goes to open a connection. One convenient way to do this is to modify compute_classpath.sh on all worker nodes to include your driver JARs. Alternatively, you can modify the compute_classpath.sh script in all worker nodes, the Spark documentation says:
If you are using the SparkNotebook, in addition to the spark properties you also need to make the driver available to the JVM that runs the back-end server.
You can do it by setting EXTRA_CLASSPATH before to start the notebook:
export EXTRA_CLASSPATH=path_to_the_first_jar:path_to_the_second_jarDataFrame Partitions
Once we have successfully set up the JDBC drivers we can now use the read.jdbc API from DataFrame to load a particular table from source. See http://spark.apache.org/docs/1.5.0/sql-programming-guide.html#jdbc-to-other-databases for documentation.
The default configuration only requires you to specify:
- url, the JDBC url to connect to (e.g. “jdbc:teradata://hostname.mycompany.com/,,”)
- dbtable, either the table name or a query wrapped between parenthesis
- driver, the driver class name (e.g. “com.teradata.jdbc.TeraDriver”)
This will create one single partition and perform a SELECT * on all of the rows and columns through a single connection. This setting might be fine if the table is small enough but is not scalable for large data.
In order to parallelize the query we have two options:
- Partitioning by uniform ranges of a specified column; or
- Partitioning by custom predicates.
Partitioning By Uniform Ranges
We need to specify extra parameters:
- columnName, the column used for partitioning. The contents of this column must be numbers.
- lowerBound, the minimum value of the column we want to select.
- upperBound, the maximum value of the column we want to select.
- numPartitions, in how many sub ranges we want to split. Please pay attention that even though Spark can distribute the collection over hundreds of partitions, the data warehouse may set some limitations on the number of parallel queries from the same user or on the allocated spool space. We suggest keeping this value low and to repartition into more folds after have been loaded from the JDBC source.
- connectionProperties, specify optional driver specific properties for tuned optimizations.
Starting from Spark 1.5.0+ :
sqlctx.read.jdbc(url = "<URL>", table = "<TABLE>",
columnName = "<INTEGRAL_COLUMN_TO_PARTITION>",
lowerBound = minValue,
upperBound = maxValue,
numPartitions = 20,
connectionProperties = new java.util.Properties()
)Partitioning By Custom Predicates
In some cases there is no uniform numerical column to partition on. In other cases we might simply want to filter the data using custom logic.
To do this, specify an array of strings where each string represents a predicate to be inserted in the WHERE statement.
For example, let’s suppose we are interested in partitioning on a specific ranges of dates, we could write it as follows:
val predicates = Array("2015-06-20" -> "2015-06-30", "2015-07-01" -> "2015-07-10", "2015-07-11" -> "2015-07-20",
"2015-07-21" -> "2015-07-31").map {
case (start, end) => s"cast(DAT_TME as date) >= date '$start' " + "AND cast(DAT_TME as date) <= date '$end'"
}
sqlctx.read.jdbc(url = "<URL>", table = "<TABLE>", predicates = predicates, connectionProperties = new java.util.Properties())This will generate a bunch of SQL queries with a WHERE statement that looks like:
WHERE cast(DAT_TME as date) >= date '2015-06-20' AND cast(DAT_TME as date) <= date '2015-06-30'See documentation at https://github.com/apache/spark/blob/v1.5.0/sql/core/src/main/scala/org/apache/spark/sql/DataFrameReader.scala
Union of Tables
Suppose now that our raw data spans over multiple tables, each with the same schema. We could first map them individually and then concatenate them into a single DataFrame using the unionAll operator:
def readTable(table: String): DataFrame
List("<TABLE1>", "<TABLE2>", "<TABLE3>").par.map(readTable).reduce(_ unionAll _)The .par is a Scala feature that simply means that the individual readTable function calls can happen in parallel rather than sequentially. The Scala framework will automatically spin one thread for each call based on the idle CPUs.
Typed Case Class Mapping
After we have constructed the DataFrame collection from the raw source we can now map it into an RDD of our ad-hoc case classes. Since a DataFrame is also an RDD of type org.apache.spark.sql.Row, it already provides the map/flatMap methods.
If there are no null values in any row, we could use pattern matching to extract each column from the Row object:
case class MyClass(a: Long, b: String, c: Int, d: String, e: String)
dataframe.map {
case Row(a: java.math.BigDecimal, b: String, c: Int, _: String, _: java.sql.Date,
e: java.sql.Date, _: java.sql.Timestamp, _: java.sql.Timestamp, _: java.math.BigDecimal,
_: String) => MyClass(a = a.longValue(), b = b, c = c, d = d.toString, e = e.toString)
}This approach will fail for null values due to the casting of the explicit types of each single field in the unapply method of the class Row. You can discard all the rows containing null values by doing:
dataframe.na.drop()But that will drop records even if the null fields are not the ones we use in our case class.
If you want to handle it using Scala options you could turn the Row object into a List and then use the following pattern:
case class MyClass(a: Long, b: String, c: Option[Int], d: String, e: String)
dataframe.map(_.toSeq.toList match {
case List(a: java.math.BigDecimal, b: String, c: Int, _: String, _: java.sql.Date,
e: java.sql.Date, _: java.sql.Timestamp, _: java.sql.Timestamp, _: java.math.BigDecimal,
_: String) => MyClass(a = a.longValue(), b = b, c = Option(c), d = d.toString, e = e.toString)
}If the columns you are interested are sparse, then you could fetch them individually either by index or by column name:
row.getAs[SQLPrimitveType](columnIndex: Int)
row.getAs[SQLPrimitveType](columnName: String)For the list of mapping of SQL primitive types and their corresponding Java/Scala classes, see: https://spark.apache.org/docs/latest/api/java/org/apache/spark/sql/Row.html.
N.B. The described procedure does not take advantage of the recently released DataSet API (http://spark.apache.org/docs/1.6.0/sql-programming-guide.html#datasets) which should automate the whole process of converting between DataFrames and RDDs. At the time we wrote this note we had not yet tested DataSet. Also there are open-source projects like Frameless (https://github.com/adelbertc/frameless) and an ongoing discussion on its gitter channel of how to leverage the awesome Shapeless (https://github.com/milessabin/shapeless) library to make Spark more functional and compile-time type-safe.
Issues With Existing Architecture
The main problem with our existing methodology is that the Spark cache is volatile across different jobs. Even though Spark provides a cache functionality, every time we restart the context, update the dependency jars or re-submit the job, the loaded data is dropped from the memory and the only way to restore it is to reload it from the central warehouse.
The following is a chart showing the time to load different tables (12 partitions each) from our data warehouse into a Spark cluster of 6 nodes in function of the row count:
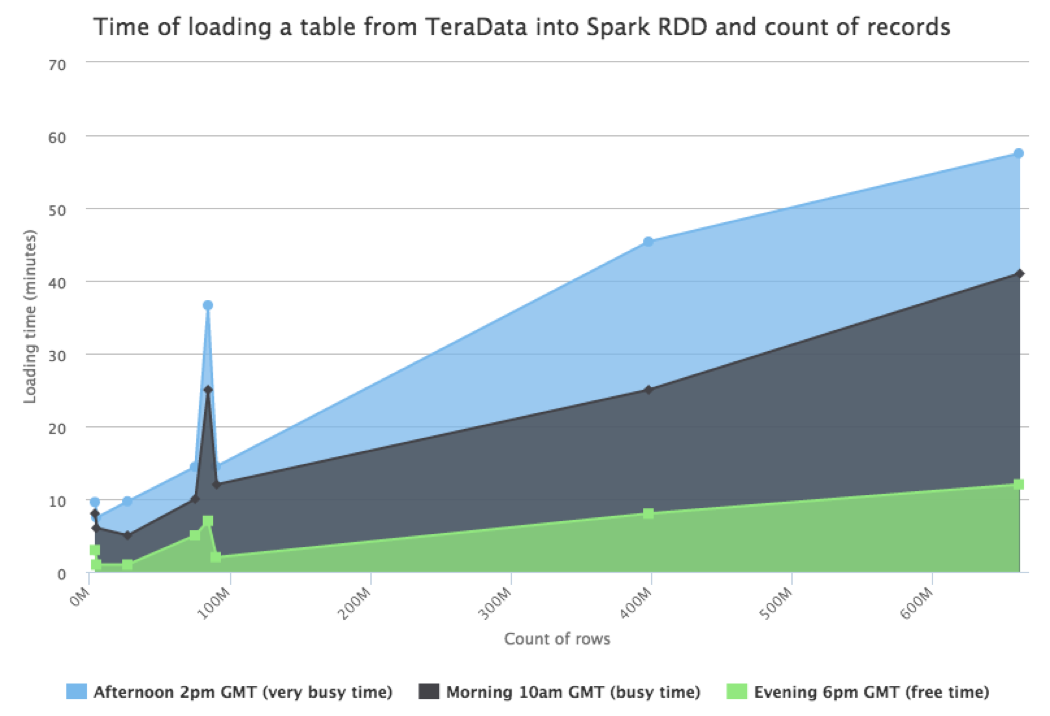
As we can see from the chart, the loading process may take minutes or hours depending on the data volume and how busy the database is. Considering that we have on average dozens of restarts per day, we cannot just rely on the Spark cache alone.
We would like to:
- Cache the raw DataFrame so we can iterate until we find the right mapping configuration.
- Cache the typed RDD for interactive exploratory analysis.
- Quickly fetch our intermediate results for responsive access and sharing the data between different applications.
In short, we want an in-memory storage system.
Tachyon as the Key Enabling Technology
Tachyon is an in-memory storage system that solves our issues and enables us to take the current deployment to the next level.
We set up Tachyon in our Spark cluster and configured no under file system (which may be Amazon S3, or HDFS, or other storage systems).
Since Tachyon is being used as an in-memory distributed file system, we can use it as storage for any text format and/or efficient data formats (such as Parquet, Avro, Kryo) together with compression algorithms (such as Snappy or LZO) to reduce the memory occupation.
To integrate with our Spark applications, we simply have to call the load/save APIs of both DataFrame and RDD and specify the path URL including the Tachyon protocol.
By having the raw data (whether it might be in Parquet-format DataFrame or Kryo-serialized Case Classes) immediately available in our Spark nodes at any time, we can now be agile and quickly iterate with our exploratory analysis and evaluation tests. We are now able to efficiently design our model and build our MVP directly from the raw source without have to face complicated and time-consuming data plumbing operations.
The following is the diagram of our workflow after deploying Tachyon and loading the data for the first time:
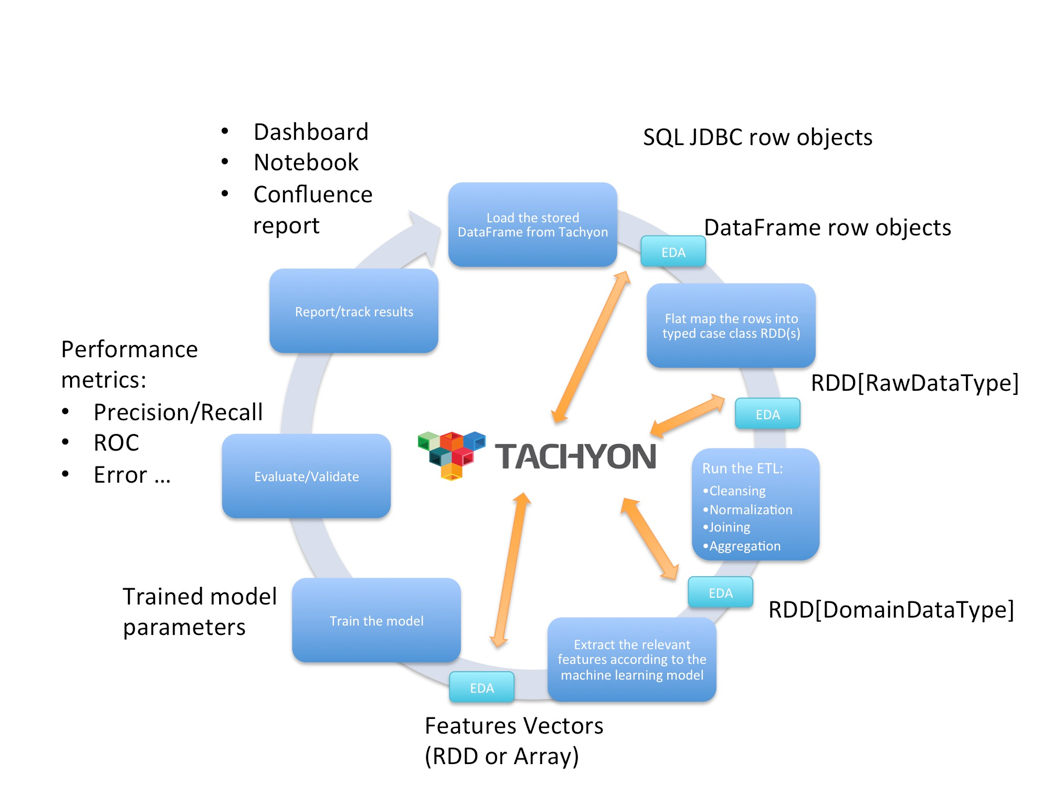
The orange arrows show stages in our workflow where the intermediate results/data are loaded/stored into Tachyon for convenient, in-memory access.
Configuring Tachyon
In our environment, we have configured Tachyon to use only the top memory tier and to work with the tmpfs mounts, typically available in unix systems under /dev/shm.
On the Tachyon master node:
1. We change the tachyon-env.sh conf file as following under the Linux settings:
export TACHYON_RAM_FOLDER="/dev/shm/ramdisk"
export TACHYON_WORKER_MEMORY_SIZE=${TACHYON_WORKER_MEMORY_SIZE:-24GB}
In the TACHYON_JAVA_OPTS we leave the default configuration with:
-Dtachyon.worker.tieredstore.level.
-Dtachyon.worker.tieredstore.level0.
-Dtachyon.worker.tieredstore.level0.dirs.path=${TACHYON_RAM_FOLDER}
-Dtachyon.worker.tieredstore.level0.dirs.quota=${TACHYON_WORKER_MEMORY_SIZE}
See http://tachyon-project.org/documentation/v0.8.2/Tiered-Storage-on-Tachyon.html
As under fs configuration we leave it empty, see https://tachyon.atlassian.net/browse/TACHYON-176.
2. We copy the new configuration to each worker:
./bin/tachyon copyDir ./conf/
3. We format Tachyon:
./bin/tachyon format
4. We deploy Tachyon without mount option (does not require root access):
./bin/tachyon-start.sh all NoMount
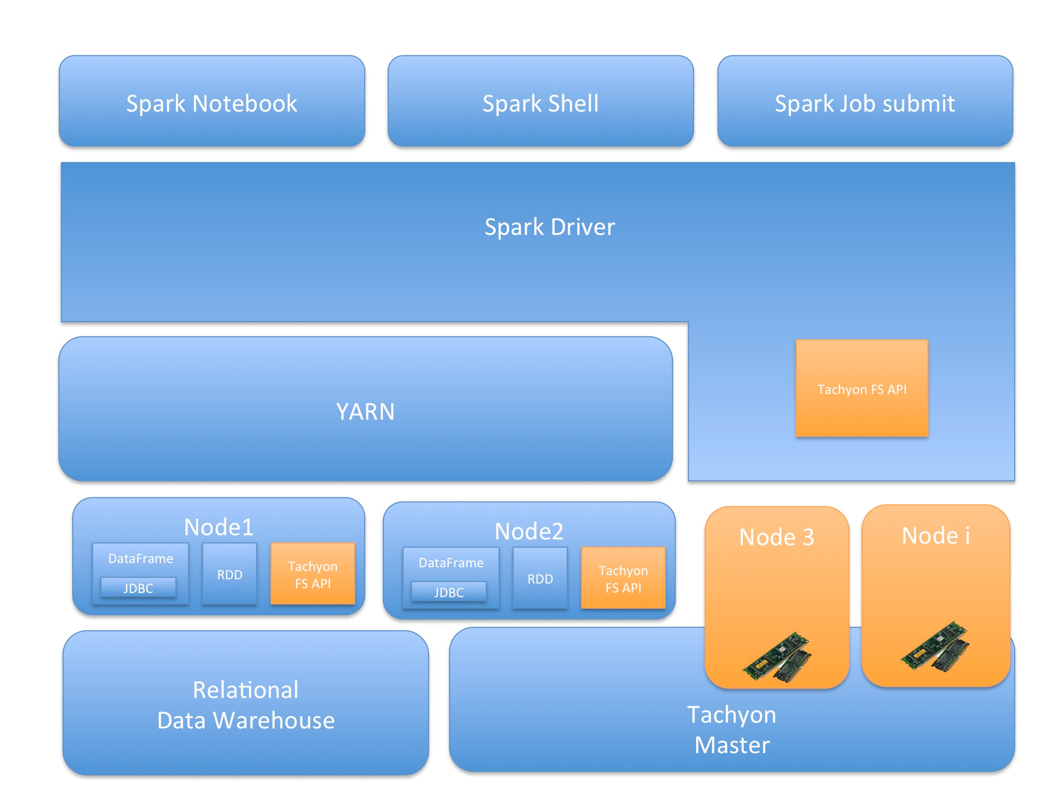
The following is the architecture diagram of all of the involved components in our configuration:
Using Tachyon With Spark
Writing and reading Spark DataFrames to and from Tachyon are very simple.
To write a DataFrame to Tachyon:
dataframe.write.save("tachyon://master_ip:port/mydata/mydataframe.parquet")Make sure that Parquet is the default format when saving in DataFrame.
To read a DataFrame from Tachyon:
val dataframe: DataFrame = sqlContext.read.load("tachyon://master_ip:port/mydata/mydataframe.parquet")See the recent documentation of Spark 1.6.0 at http://spark.apache.org/docs/1.6.0/sql-programming-guide.html#generic-loadsave-functions
Similarly, writing and reading Spark RDDs to and from Tachyon are very simple.
To write an RDD to Tachyon:
rdd.saveAsObjectFile("tachyon://master_ip:port/mydata/myrdd.object")Make sure that the default serializer is Kryo for RDDs.
To read an RDD from Tachyon:
val rdd: RDD[MyCaseClass] = sc.objectFile[MyCaseClass] ("tachyon://master_ip:port/mydata/myrdd.object")Make sure that MyCaseClass is the same class as when the RDD was cached into Tachyon, otherwise it will throw a class version error.
Making the Impossible Possible
We sped up our Agile Data Science workflow by combining Spark, Scala, DataFrame, JDBC, Parquet, Kryo and Tachyon to create a scalable, in-memory, reactive stack to explore the data and develop high quality implementations that can then be deployed straight into production.
Thanks to Tachyon, we now have the raw data immediately available at every iteration and we can skip the costs of loading in terms of time waiting, network traffic, and RDBMS activity. Moreover, after the first ETL, we save the normalized and cleaned data in memory, so that the machine learning jobs can start immediately, allowing us to run many more iterations per day.
By configuring Tachyon to keep data only in memory, the I/O cost of loading and storing into Tachyon is on the order of seconds, which in our workflow scale is simply negligible.
Our workflow iteration time decreased from hours to seconds. Tachyon enabled something that was impossible before.
Future Developments
The presented methodology is still an experimental workflow. Currently, there are some limitations and room for improvement. Here, we present some of the limitations and potential for future work.
- Since we have not specified an under file system layer, Tachyon can only handle as much data as it fits into the allocated space. The tiered storage feature in Tachyon can solve this issue.
- Setting up JDBC drivers, the partitioning strategy and the case class mapping is a big overhead and is not very user-friendly.
- Memory resources are shared with Spark and Tachyon, so to avoid duplication and unnecessary garbage collection, some fine-tuning is required
- If a Tachyon worker fails, the data is lost, since we configured no tiered storage nor under file system, and Tachyon does not know how to re-load the data from a JDBC source. This is fine for our use case, since our goal is in-memory storage layer to use as long-term cache, but it can be a very important improvement for future developments.
Nevertheless, we believe that the described methodology, combined with Tachyon, is a game-changer for effectively applying agile data science into large corporations.
***
About the authors:
Gianmario Spacagna is a Data Scientist at Barclays Personal and Corporate Bank, building scalable machine learning and data-driven applications in Spark and Scala.
Harry Powell leads the Data Science team at Barclays Personal and Corporate Bank.

